Reduced Representation Bisulfite Sequencing (RRBS)
Product Introduction
Reduced Representation Bisulfite Sequencing (RRBS) is an enzymatic-enrichment strategy that selectively captures promoter regions and CpG islands followed by bisulfite conversion and sequencing [3–5]. Compared with whole-genome bisulfite sequencing (WGBS), RRBS offers higher cost-effectiveness and requires markedly lower sequencing depth, making it well-suited for large-scale clinical studies.
Application
1.Mapping the whole genome methylation map;
2.To study the epigenetic mechanisms of physiological activities such as embryonic development and aging;
3.Study disease-related methylation molecular markers.
Advantages
1.High performance cost ratio;
2.The reduced sequencing volume is suitable for large clinical samples.
Sample Requirements
Sample Types
1.DNA≥ 1 ug/sample, concentration ≥25 ng/μL;
2.Whole blood, 1-4 mL/sample;
3.Cells ≥ 5×10^6 cells/sample;
4.Animal tissues ≥ 30 mg/sample.
Sample Species
Limited to human, mouse, and rat DNA, whole blood, Cells, animal tissues; other species require evaluation.
Analysis Content
1.Genome Alignment Statistics;
2.Quality Control of MSPI Digestion and Bisulfite Conversion Rate;
3.Methylation Site Calling;
4.Methylation Distribution Profiles;
5.Annotation of Differentially Methylated Regions (DMRs);
6.GO and KEGG Enrichment Analysis of DMR-Associated Genes.
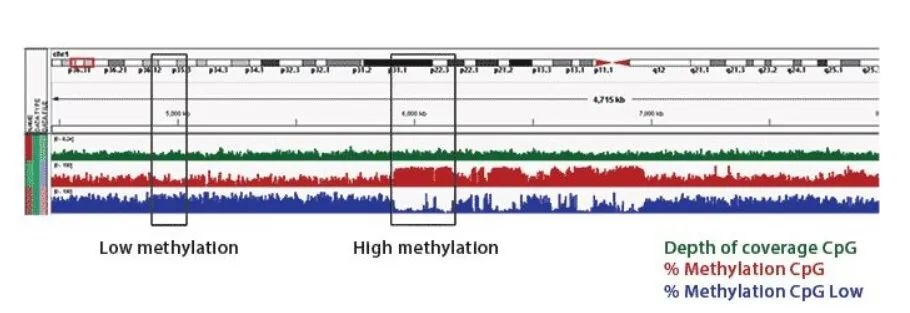
Figure 1. IGV plots of DMRs
Example results
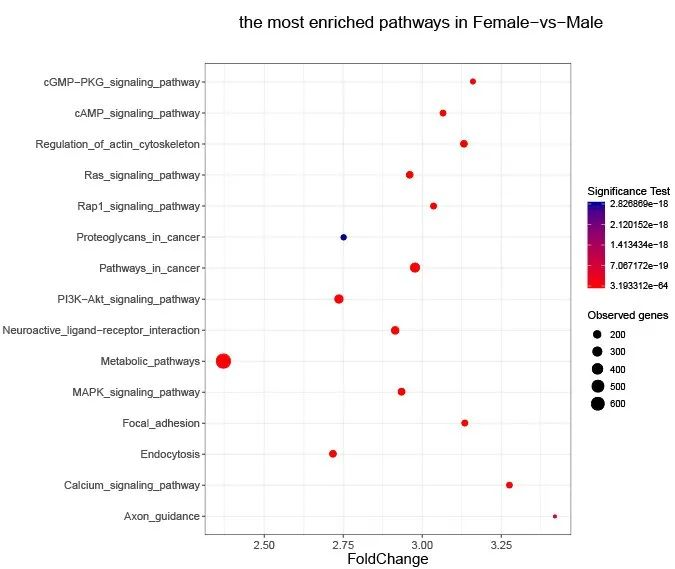
Figure 2. KEGG Enrichment Analysis of DMRs
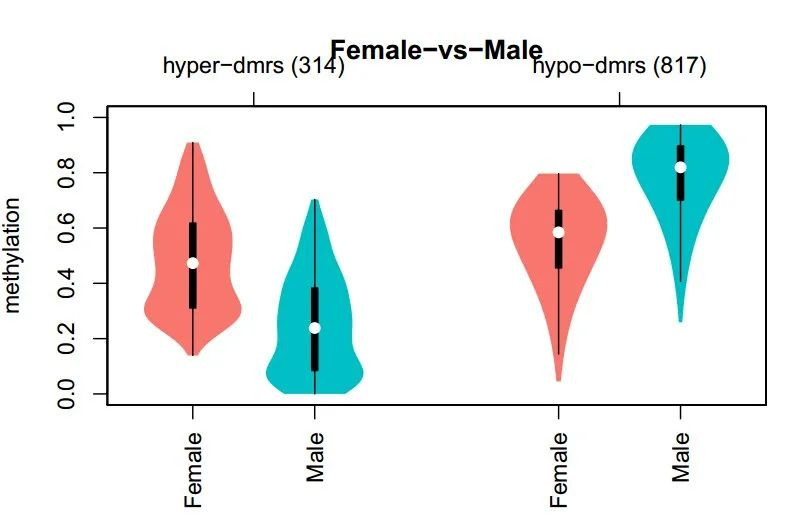
Figure 3. Average Methylation Level Analysis of DMRs
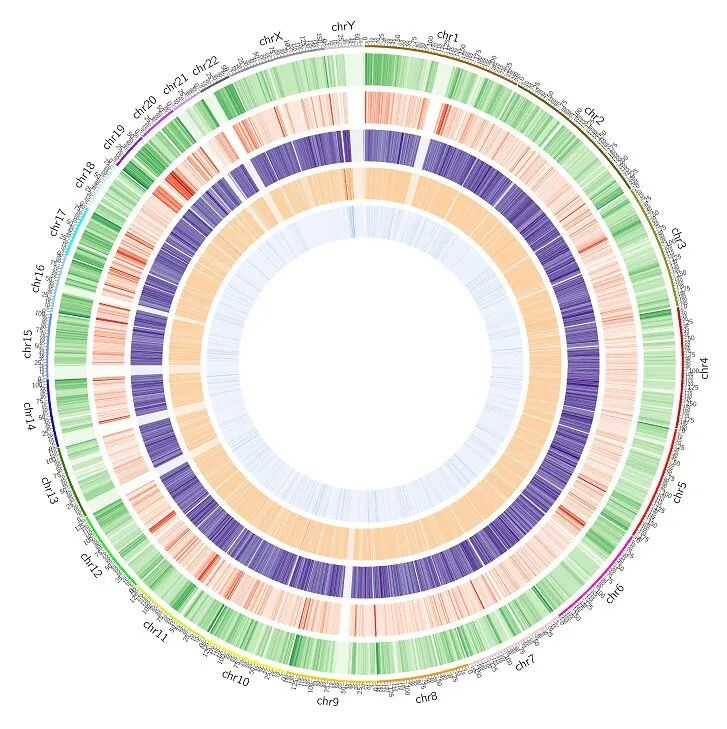
Figure 4. Chromosomal level methylation analysis
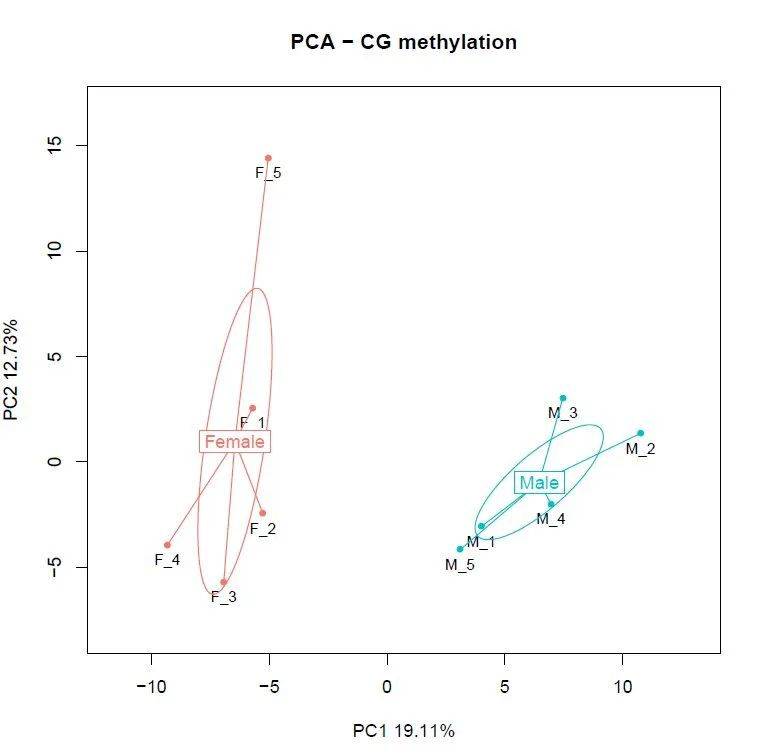
Figure 5. PCA analysis
References
[1] MeissnerA, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33(18).
[2] Frank Jühling et al. metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Research, 2016, 26: 256-262.
[3] Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 2009 Apr;27(4).
