Targeted Bisulfite Sequencing (TBS)

Product Introduction
Targeted Bisulfite Sequencing (TBS) is a DNA methylation profiling technology that combines multiplex bisulfite-PCR capture with next-generation sequencing at high depth [1-2]. By selectively interrogating predefined genes or CpG loci, TBS delivers accurate, locus-specific methylation data and has been validated for diverse clinical specimens, including blood, tissue, and bronchoalveolar lavage fluid.
Application
1.Draw the methylation map of target genes/CpG sites;
2.Screening for methylation disease markers;
3.Verify the methylation of downstream target genes.
Advantages
1.Low cost, short cycle and high cost performance;
2.High detection sensitivity, high detection flux and single base resolution;
3.Wide range of applications, covering a variety of cfDNA source samples.
Sample Requirements
Sample Types
1. DNA ≥200ng/sample, concentration ≥10ng/μL;
2. Cells, whole blood, animal tissues, etc., inquire for details.
Sample Species
Limited to human, mouse, and rat ; other species require evaluation.
Analysis Content
Basic Analysis
1.Raw reads trimming and quality control
2.Alignment to the target regional sequences
3.Statistical analysis of CpG site methylation levels
4.Comparison of methylation levels between different samples
Advanced analysis
1.clinical pathological information analysis
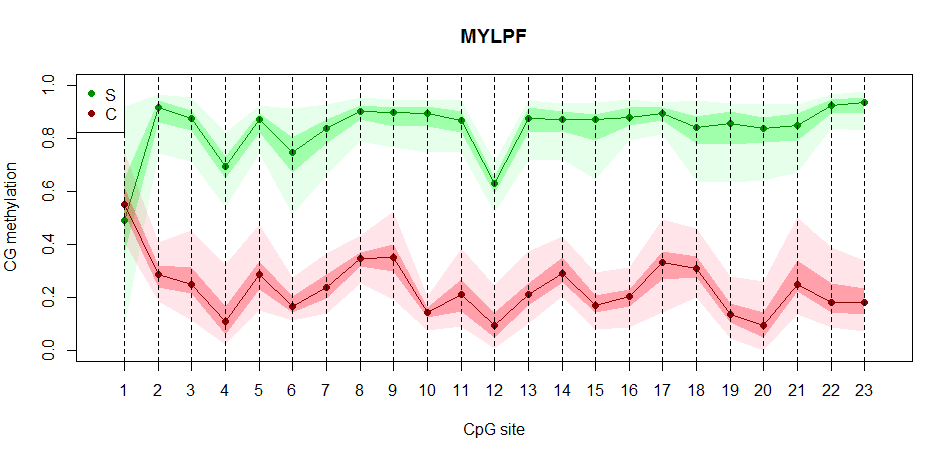
Figure 1. Schematic diagram of methylation level distribution of CG sites in the target region
Example results
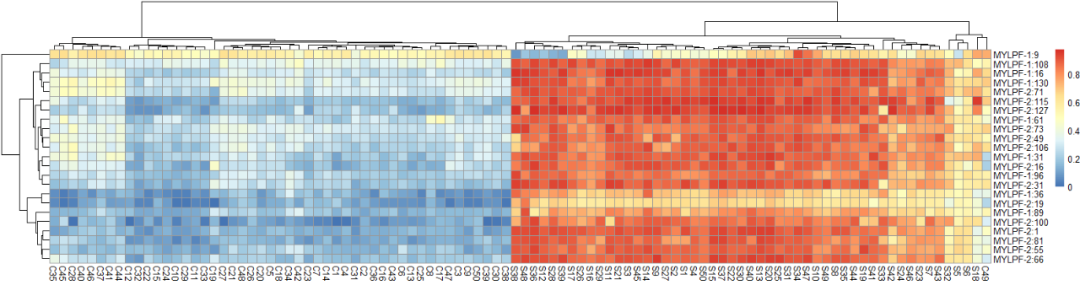
Figure 2. Multimodal methylation level clustering heat map
References
[1]Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 2009 Apr;27(4).
[2]HaiderZ, Landfors M, Golovleva I, et al. DNA methylation and copy number variation profiling of T-cell lymphoblastic leukemia and lymphoma. Blood Cancer J 2020 Apr 28;10(4)
