WGBS(Whole Genome Bisulfite Sequencing)
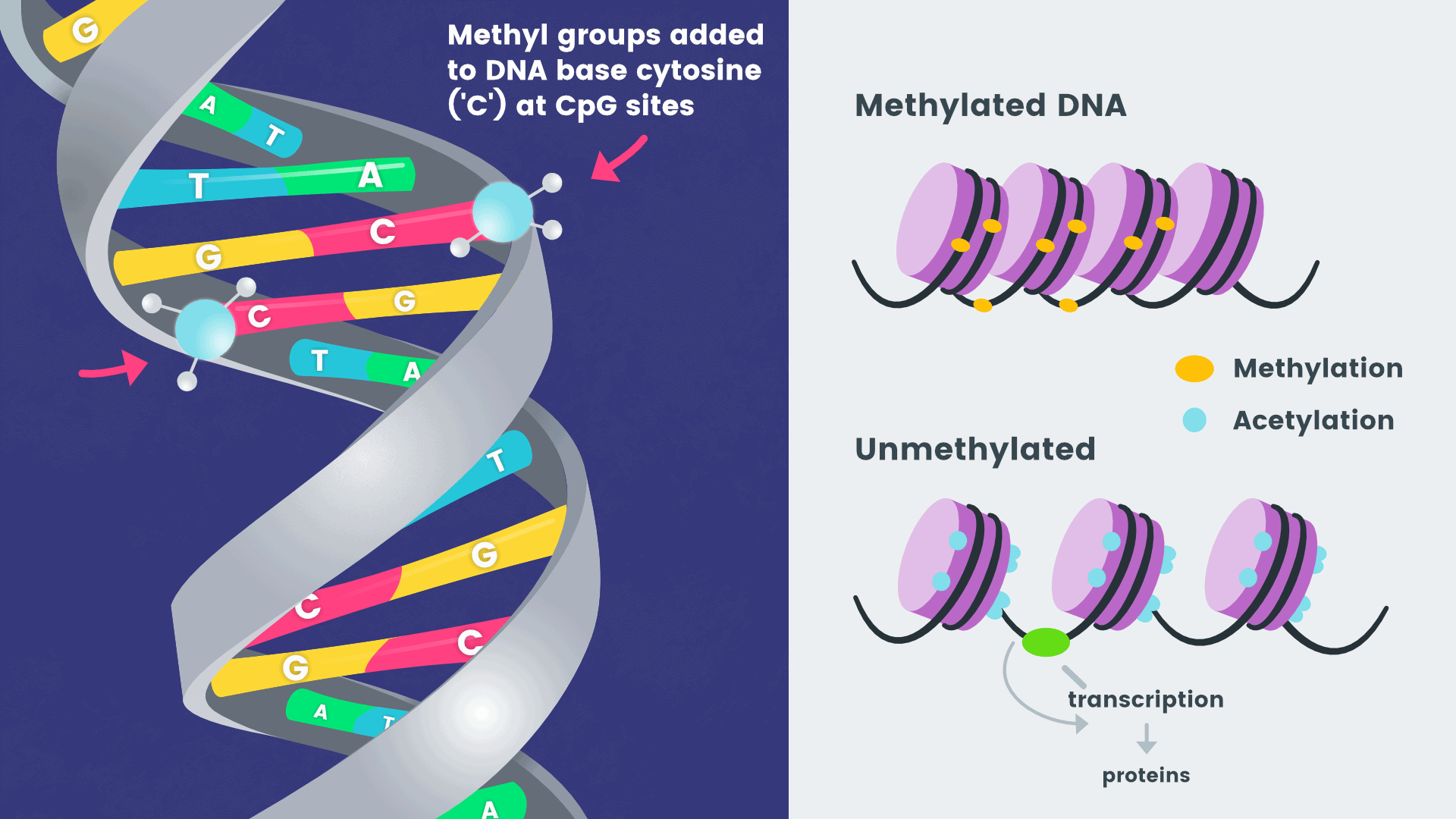
Product Introduction
Whole Genome Bisulfite Sequencing (WGBS) is the gold standard for DNA methylation research [1-2]. It employs bisulfite treatment to map the DNA methylation profile of the genome-wide with single-base resolution.
Application
1.Mapping whole genome single base resolution methylation map;
2.To study the epigenetic mechanisms of physiological activities such as embryonic development and aging;
3.Study disease-related methylation molecular markers.
Advantages
1.Precise identification of CpG enrichment sites with single base resolution
2.The whole genome coverage and the more complete methylation information obtained are the gold standard of DNA methylation research
Sample Requirements
Sample Types
1. DNA ≥3 ug/sample, concentration ≥30 ng/μL;
2. Plasma/Serum, 1~4 mL/sample;
3. cfDNA ≥15 ng.;
4. Cells, whole blood, animal tissues, etc., inquire for details.
Sample Species
Limited to human, mouse, and rat DNA, plasma/serum, cfDNA; other species require evaluation.
Analysis Content
1. Genomic alignment statistics
2. Global methylation level assessment
2.1 Average methylation level
2.2 Methylation distribution plot
3. Methylation site statistics and inter-sample comparison
3.1 Correlation analysis of methylation levels
3.2 Principal component analysis (PCA) of methylation levels
3.3 Cluster analysis of methylation levels
4. Differential methylation analysis
4.1 Annotation of differentially methylated regions (DMRs)
4.2 Length distribution of differentially methylated regions
4.3 Methylation level distribution of DMRs
4.4 Cluster heat map of methylation levels in DMRs
4.5 Visualization of DMRs
4.6 GO and KEGG enrichment analysis of genes related to DMRs
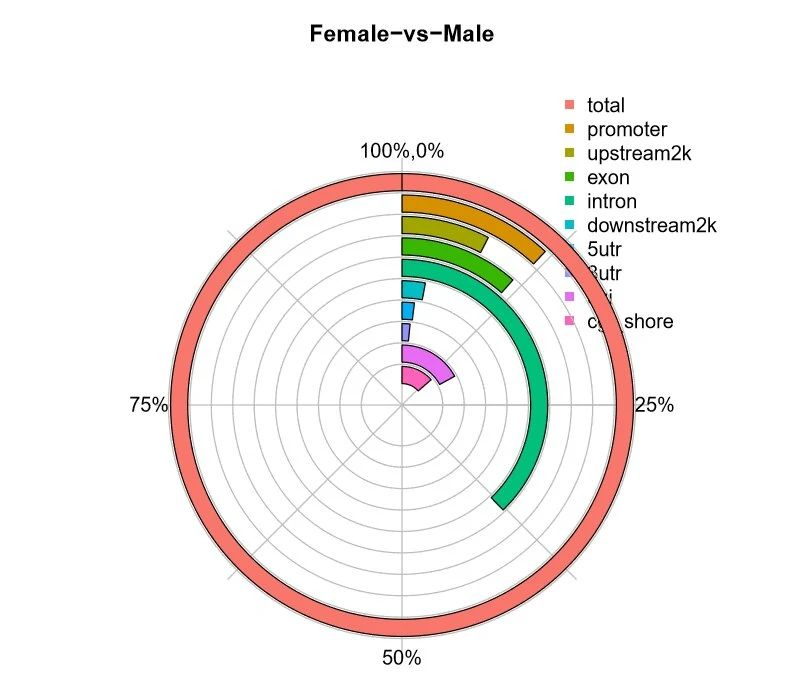
Figure 1. Proportion of DMR distribution in the number of anchoring regions
Example results
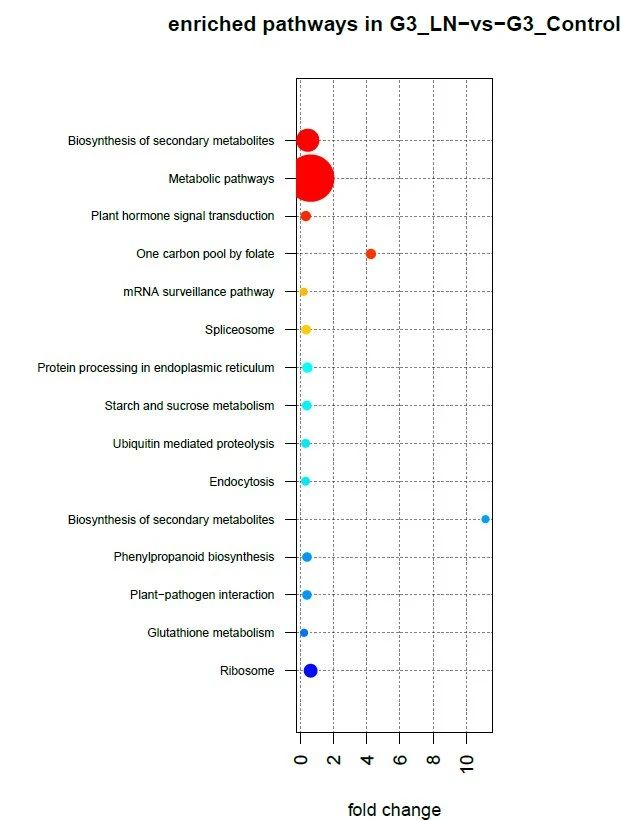
Figure 2. KEGG analysis of genes related to DMR anchored promoter region
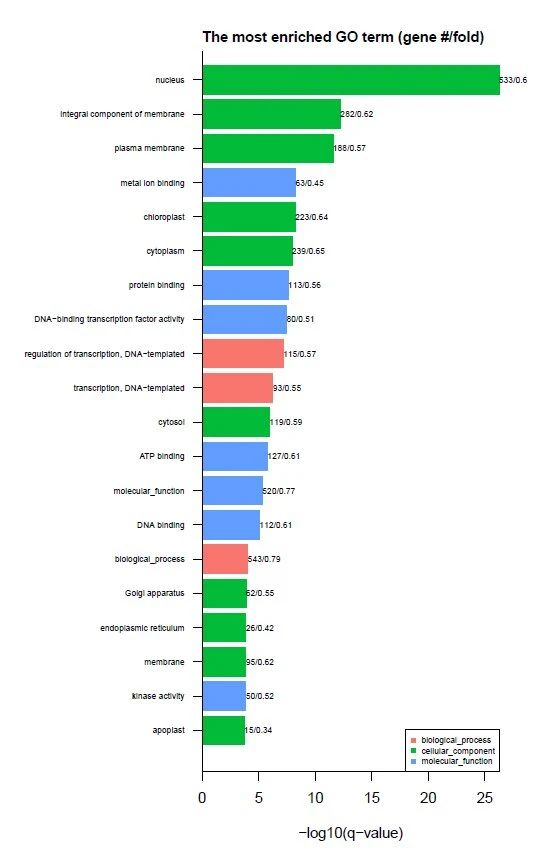
Figure 3. GO enrichment analysis of DMR related genes
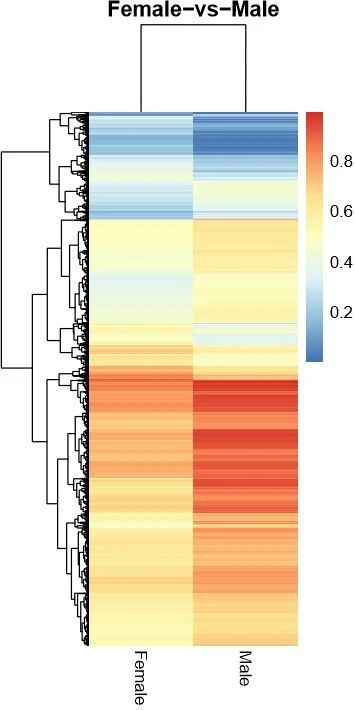
Figure 4. Cluster heat map of methylation levels in differentially methylated regions
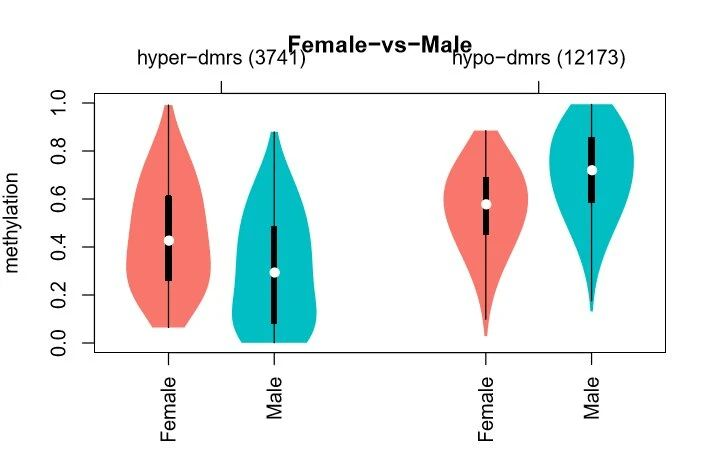
Figure 5. Distribution of average methylation levels in differentially methylated regions
References
[1] Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature.2014.
[2] Ehsan Habibi, et al. Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells. Cell Stem Cell (2013), 13(3), 360-369.
